Get Started
Contents
Get Started#
Install#
Precondition: You need a Linux Computer with an already installed Anaconda or Miniconda environment.
Download ZIP-file from XXX, store it to an arbitrary work directory, say /home/dsci/b/gendifs/. Unzip the zip file. Among other directories you will get the subdirs mm, md, py.
Start Juypter. In the dasci-lab this will work:
Change into the subdir py of your work directory. Among other files you should be able to detect the python program gd05.py.
Create your first GenDifS mindmap#
We show a full cycle of how to craft a simple taxonomical ontology with GenDifS.
Say you want to formalize the example from the SKOS-Primer, Section 4.1:
milk
<milk by source animal>
cow milk
goat milk
Start freeplane. Create a new empty mindmap. Select (CTRL-C) the following lines:
ONTOLOGY
milk
BY SOME mammal
cow milk
SOME cow
goat milk
SOME goat
Paste (CTRL-V) it to the root node of the mindmap. You should get a mindmap like this:
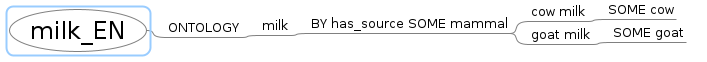
Save this mindmap to the mm directory within your work directory, e.g. /home/dsci/b/gendifs/mm/milk_EN.mm.
(Of course we already have allocated this mindmap for you in the mm directory, see ../mm/milk_EN_initial.mm. You may open it, and save it again to milk_EN.mm.)
Use gd05.py#
Go back to Juypter. Navigate to the directory py within your work directory (in our example /home/dsci/b/gendifs/py).
Create an new Python 3.x Jupyter Notebook, say my_sandbox.ipynb.
from gd05 import GenDifS_Map
import os
Create an instance of Class Ontology:
mm_relative = "../mm/milk_EN.mm"
# provide an absolute path to get well definded file locations
mm = os.path.abspath(mm_relative)
o = GenDifS_Map(mm)
cwd into input_dir: /home/dsci/a/l/LA_2022_ws/mm
reading /home/dsci/a/l/LA_2022_ws/mm > milk_EN.mm (8 nodes)
Parsing XML: #1 start nodes
ontology node IDs: {'ID_397914510': 'ONTOLOGY'}
RDF: 102 498
warning: no glossary file found.
updated mindmap with backup to /home/dsci/a/l/LA_2022_ws/mm/milk_EN.mm_backup_2022-11-20T13-49-53
The parser will read the mindmap file, parse it’s content, and generate several internal and external representations of the ontology.
External Representations (i.e. files)#
As a first step, feedback is provided within the mindmip itself. Warning: This feedback will overwrite your file milk_EN.mm (and allocate a backup file directly in the mm-directory). To see the changes, switch back to freemind, and reload the mindmap:
Datei > aus Revisionsverlauf wiederherstellen > “milk_EN.mm” > Öffnen
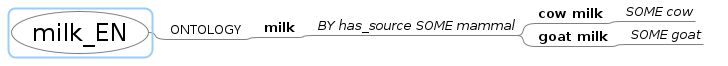
The syntax highlight will visualize classes, other stuff, and also Errors.
o = GenDifS_Map(mm, write_ontology = True)
cwd into input_dir: /home/dsci/a/l/LA_2022_ws/mm
reading /home/dsci/a/l/LA_2022_ws/mm > milk_EN.mm (8 nodes)
Parsing XML: #1 start nodes
ontology node IDs: {'ID_397914510': 'ONTOLOGY'}
RDF: 102 498
warning: no glossary file found.
updated mindmap with backup to /home/dsci/a/l/LA_2022_ws/mm/milk_EN.mm_backup_2022-11-20T13-49-53
writing ontologies to /home/dsci/a/l/LA_2022_ws/milk_EN.mm_ONTOLOGY/ :
gendifs.ttl rdflib.ttl
owlrl.ttl
You also will get 3 representations of your ontology:
../milk_EN.mm_ONTOLOGY/gendifs.ttl: the raw Turle code produced by GenDifS itself../milk_EN.mm_ONTOLOGY/rdflib.ttl: the turtle code exported by rdflib../milk_EN.mm_ONTOLOGY/owlrl.ttl: the turtle code exported by owlrl after applying a full deductive closure inferencing
Each of these ttl-files load into Protegé directly. However, they look somewhat crowded:
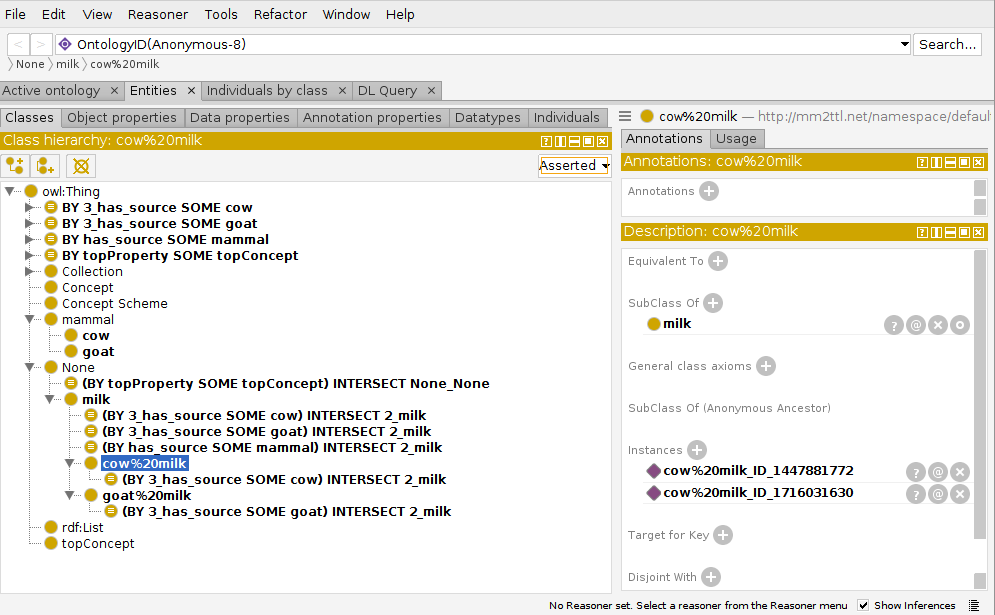
Internal Representations#
The external representations correspond with internal representations:
o.ttl: The raw GenDifS ttl, given as a stringo.rdflib: A rdflib-Graph, as it is after importing the raw GenDifS-ttlo.owlrl: The rdflib-Graph, after applying owlrl-inferencing.
print(o.ttl[0:300]) # GenDifS simply exports to Turtle (*.ttl), i.e. a human readable text file
# namespaces to be aligned by the user
@prefix ex: <http://mm2ttl.net/namespace/ex#> . # examples of OWL A-Box
@prefix cpt: <http://mm2ttl.net/namespace/cpt#> . # concept; instance of skos:concept
@prefix : <http://mm2ttl.net/namespace/default#> .
print(f"{type(o.rdflib)}: {len(o.rdflib)} triples") # a more complex and powerful python object
<class 'rdflib.graph.Graph'>: 102 triples
rdflib allows to query it’s graphs via SPARQL:
my_query = """
SELECT DISTINCT ?s ?p ?o
WHERE {
?s ?p ?o .
?s rdf:type :milk
}"""
for row in o.rdflib.query(my_query):
print(row.s, row.p, row.o)
http://mm2ttl.net/namespace/ex#milk_ID_564335606 http://www.w3.org/1999/02/22-rdf-syntax-ns#type http://mm2ttl.net/namespace/default#milk
http://mm2ttl.net/namespace/ex#milk_ID_1716031630 http://www.w3.org/1999/02/22-rdf-syntax-ns#type http://mm2ttl.net/namespace/default#milk
http://mm2ttl.net/namespace/ex#milk_ID_1716031630 http://mm2ttl.net/namespace/default#has_source http://mm2ttl.net/namespace/ex#cow_ID_1716031630
http://mm2ttl.net/namespace/ex#milk_ID_1716031630 http://mm2ttl.net/namespace/ex#classifyLike http://mm2ttl.net/namespace/ex#cow%20milk_ID_1716031630
http://mm2ttl.net/namespace/ex#milk_ID_1490639552 http://www.w3.org/1999/02/22-rdf-syntax-ns#type http://mm2ttl.net/namespace/default#milk
http://mm2ttl.net/namespace/ex#milk_ID_1490639552 http://mm2ttl.net/namespace/default#has_source http://mm2ttl.net/namespace/ex#goat_ID_1490639552
http://mm2ttl.net/namespace/ex#milk_ID_1490639552 http://mm2ttl.net/namespace/ex#classifyLike http://mm2ttl.net/namespace/ex#goat%20milk_ID_1490639552
print(f"{type(o.owlrl)}: {len(o.owlrl)} triples")
<class 'rdflib.graph.Graph'>: 498 triples
